Overview
Many genebanks globally contain untapped resources of distinct alleles which will remain hidden unless efforts are initiated to screen these alleles of its potential use and function.
The deployment of useful diversity using core collections has been an area of much interest for researchers especially those working in the field of allele mining. The prerequisite of any core collection established is that it captures the complete diversity of the entire collection it was derived from. A core set should not be considered a substitute of the entire collection.
The recent advancements in technological tools related to genomics and bioinformatics have made it possible to discover new alleles for any gene of interest. These new techniques also create a further challenge of linking traditional phenotypic information to a larger quantity of sequential and genetic information and to complement activities carried out for germplasm enhancement. Allele mining provides the avenue for the validation of specific gene (s) responsible for a particular trait and mining of the most favorable alleles.
The advent of PowerCore that implements the advance M-strategy using a modified heuristic algorithm (A*) is hoped to provide users the added ability to develop core or allele mining sets representing all alleles or classes of their observations whilst ensuring the least allelic redundancy and highly reproducible list of entries..
PowerCore software uses the .NET Framework Version 1.1 environment and is freely available for the MS Windows platform.
The recent advancements in technological tools related to genomics and bioinformatics have made it possible to discover new alleles for any gene of interest. These new techniques also create a further challenge of linking traditional phenotypic information to a larger quantity of sequential and genetic information and to complement activities carried out for germplasm enhancement. Allele mining provides the avenue for the validation of specific gene (s) responsible for a particular trait and mining of the most favorable alleles.
The advent of PowerCore that implements the advance M-strategy using a modified heuristic algorithm (A*) is hoped to provide users the added ability to develop core or allele mining sets representing all alleles or classes of their observations whilst ensuring the least allelic redundancy and highly reproducible list of entries..
PowerCore software uses the .NET Framework Version 1.1 environment and is freely available for the MS Windows platform.
Screenshots
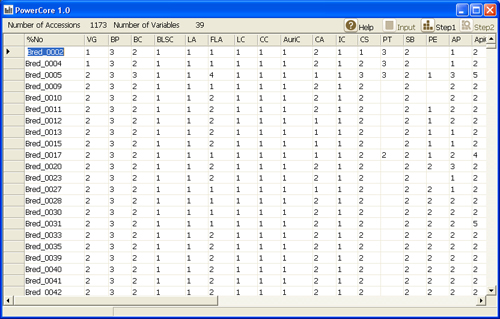
Directly attached from Excel worksheet
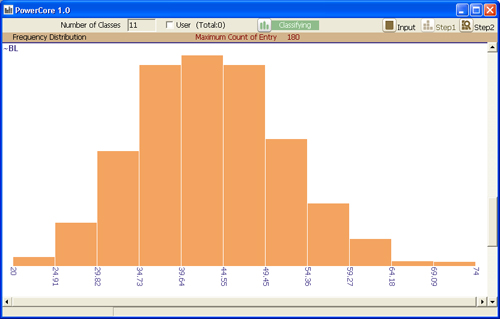
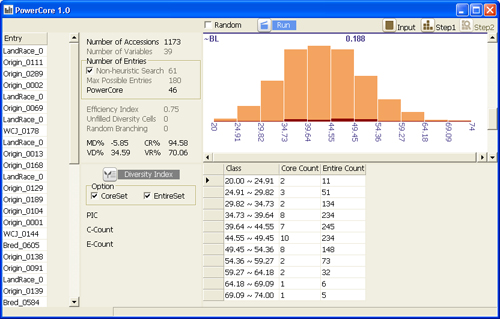
Automatically classified diversity distribution of accessions
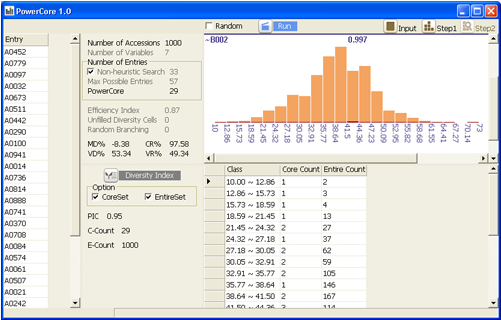
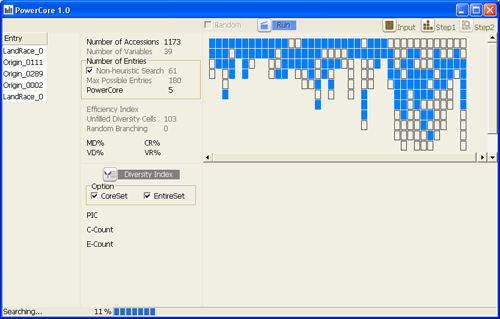
Selecting core set by heuristic search based on modified A* algorithm
Displaying results
Downloads
Software
| Name | Descriptions | Release Date |
|---|---|---|
| PowerCore_Software_V1.zip |
PowerCore software version 1.0 for installation |
2/22/2007 |
| PowerCore_Software_V2.zip |
PowerCore software version 2.0 for Windows 10 |
11/4/2021 |
Manual
| Name | Descriptions | Release Date |
|---|---|---|
| PowerCore_User_Manual.pdf | PowerCore user manual | 2/22/2007 |
| Bioinformatics Publication | 6/22/2007 |
Supplementary data
- i. SSR and phenotypic data sets for 1000 accessions of rice
| Name | Descriptions | Release Date |
|---|---|---|
| Phenotypic_Dataset_for_PowerCore.xls | PowerCore readable phenotype excel data of rice accessions | 2/22/2007 |
| SSR_Dataset_for_PowerCore.xls | PowerCore readable SSR excel data of rice accessions | 2/22/2007 |
| Results_for_Phenotypic_Dataset.xls | PowerCore-phenotypic output data | 2/22/2007 |
| Results_for_SSR_Dataset.xls | PowerCore-SSR output data | 2/22/2007 |
- Virtual 1000 accessions
| Name | Descriptions | Release Date |
|---|---|---|
| Virtual_1000.xls | Original 1000 virtual accessions | 2/22/2007 |
| Virtual_1000_for_PowerCore.xls | Converted format acceptable by PowerCore | 2/22/2007 |
| Virtual_1000_for_MSTRAT.zip | Converted format acceptable by MSTRAT | 2/22/2007 |
| Results_for_Virtual_1000.xls | Results of 1000 virtual data by PowerCore and MSTRAT | 2/22/2007 |

